Examples
# Good
fit_model <- function() {
# code
}
results_df <- data.frame()At ESQlabs, we follow the tidyverse style guide with very few changes. The following sections outline the most important differences with the Tidyverse guide. To learn more about original Tidyverse style guide, please refer to the Tidyverse style highlights section.
For simplicity, users should use snake_case as recommended by the tidyverse style (this is not the case for developers that should follow the OSPS-R coding standards).
However the naming style depends on the type of object:
Variable and function names should use only letters and numbers. Use snake_case to separate words within a name:
True constant variables should use ALL_CAPS casing:
Use short meaningful and understandable names. The code should read as a story and only some well-known abbreviations should be used:
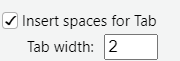
Because it is not explicitly said in the tidyverse guideline and for improved code readability, use the following indentation settings:
You can check that these settings are correctly set in Tools > Global Options > Code > Editing:
return()Prefer using return() for explicitly returning result, although you can rely on R to implicitly return the result of the last evaluated expression in a function.
Avoid having long lines. Restrict yourself to 120 characters (instead of the usual limit of 80 characters).
The tidyverse style guide is the reference regarding R coding style. The guide is quite comprehensive, with a lot of examples of what to do and what not to do. If you never read it, you should read it at least once.
Here we highlight the main aspects of the tidyverse style guide, in a very synthetic way. This is to be used more as a checklist. Please refer to the full tidyverse style guide for more details and examples.
Examples listed here might have been adapted, when appropriate, to follow ESQlabs’ style instead of the Tidyverse one.
.R.-, and _.- and =) to break up your file into easily readable chunks._ to separate words within a name (so called snake case).<-, not =, for assignment.+, -, /, *, <-, =, ==, &, &&, |, ||, >, <, >=, <=, %in%, etc.) should always be surrounded by spaces.^, :, $, [], [[]], ::, :::, @, etc.) should never be surrounded by spaces.<- or =.= when used in function calls.() used for function arguments definition.() when used with if, for, or while.else should be on the same line as }.& and | should never be used inside of an if clause because as they can return vectors. Always use && and || instead.{ should always be followed by a new line. Related code (e.g., an if clause, a function declaration, …) must be on the same line as the opening brace.} should always go on its own line, unless it’s followed by else.%>% should always have a space before it, and should usually be followed by a new line.+ should always have a space before it, and should be followed by a new line (such that each layer is on its own line).). This makes the code easier to read and to change later.( of the function). This is the preferred method.) and leading { should go on the same line as the last argument.# Function indent (preferred method)
long_function_name <- function(a = "a long argument",
b = "another argument",
c = "another long argument") {
# As usual code is indented by two spaces
}
# Double indent
long_function_name <- function(
a = "a long argument",
b = "another argument",
c = "another long argument") {
# As usual code is indented by two spaces
}TRUE and FALSE over T and F.", not ', for quoting text.; at the end of a line, and don’t use ; to put multiple commands on one line.
Comments
#and a single space.